On February 3, the Wuhan virus research institute of the Chinese academy of sciences/biosafety research center, Wuhan jinyintan hospital and Hubei provincial center for disease prevention and control cooperated in the identification and research of 2019 new coronavirus (2019-nCoV), which caused the Wuhan pneumonia epidemic, and published it online under the topic "a pneumonia outbreak associated with a new coronavirus of disease origin" (the link of the paper is https://www.nature.com/articles/s41586-020-2012-7). Relevant research teams revealed the basic biological characteristics of the coronavirus from nucleic acid detection, serological diagnosis, virus isolation and receptor utilization, providing important clues for epidemic control and drug research and development.
The research team obtained the whole genome sequence of the virus from the early samples of 5 patients. The similarity of the virus sequence from 5 patients reached 99.9%, and the sequence consistency with SARS-CoV was 79.5%. The amino acid analysis of virus conserved protein shows that 2019-nCoV and SARS-CoV belong to SARS-related coronavirus (SARSR-COV). The author further compared 2019-nCoV genome with some gene sequences of coronavirus detected early in the laboratory, and found that the virus is similar to the gene of a coronavirus (TG13 for short) derived from a bat sample. After sequencing the bat sample, the whole genome sequence of bat virus TG13 was obtained, and the sequence consistency of the two viruses was found as high as 96% (Figure 1).
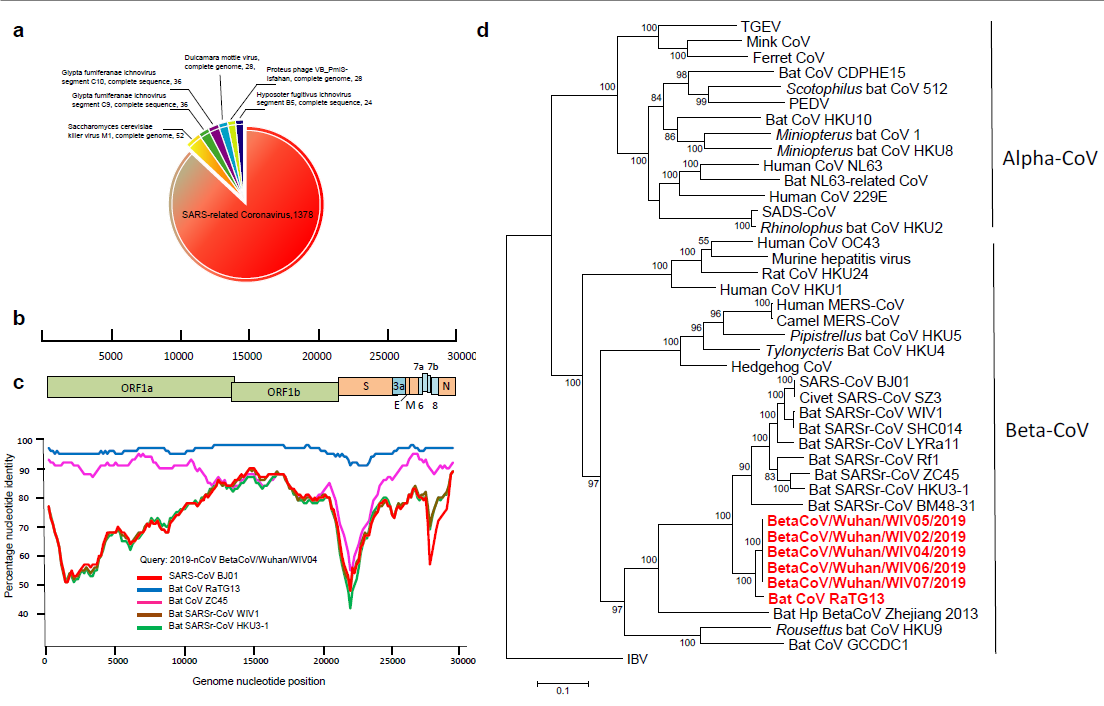
Fig. 1. high-throughput sequencing obtains the whole genome sequence (A) of the virus from a patient sample; Sketch map of 2019-nCoV genome (b); Homology analysis with human SARS virus and bat SARSr-CoV (c); Evolutionary analysis using 2019-nCoV replicase gene (d)
The research team carried out nucleic acid detection on samples of bronchoalveolar lavage fluid (BALF), throat swab (OS) and other samples of patients in different periods, and found that the viral nucleic acid of early samples was about 1000 times higher than that of late samples. By using SARSr-CoV antigen stored in the laboratory at an early stage, they tested the patient's serum samples and found that the patient produced corresponding IgM and IgG antibodies (fig. 2). Subsequently, 2019-nCoV virus was isolated from bronchoalveolar lavage fluid of a critically ill patient, and its intracellularly clear coronavirus particle morphology was observed by electron microscopy (fig. 3).
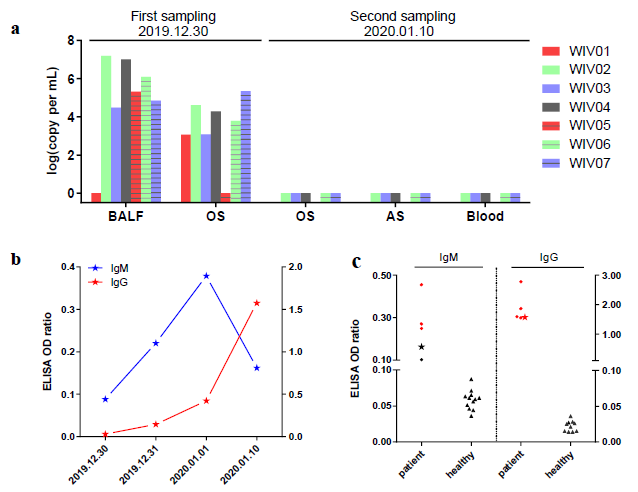
Fig. 2. virus nucleic acid detection in seven patients (a); Serological changes in a patient (B); Detection of Antibody Level in Seven Patients (C)
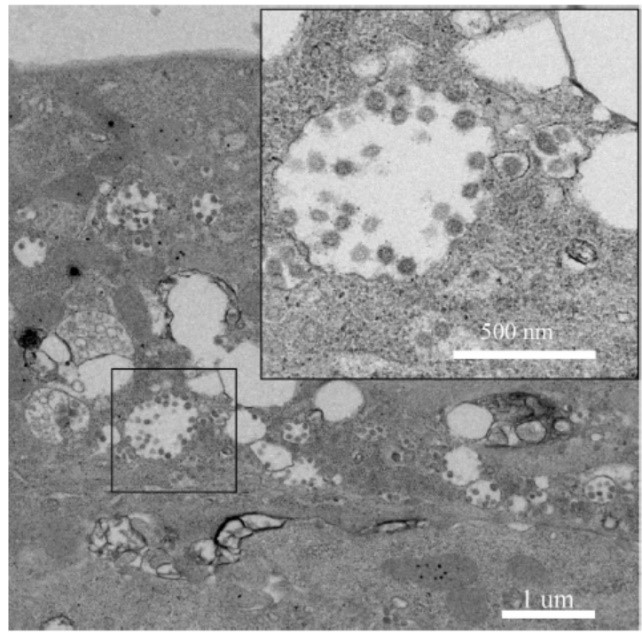
Fig. 3. electron microscopic observation of 2019-ncov in cells
The research team found that 2019-nCoV can infect non-sensitive cells that express human ACE2 (angiotensin converting enzyme, cell receptor of SARS-CoV), indicating that 2019-nCoV can invade cells using receptor of SARS-CoV (fig. 4).
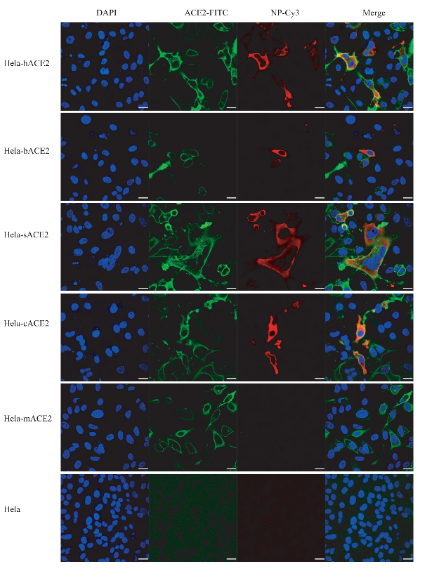
Fig. 4. 2019-nCoV infection in HeLa cells expressing ACE2 from human, bat, pig, civet and mouse
In the race against time and epidemic situation, the study provided more detailed basic biological characteristics of 2019-nCoV virus in a short period of time, and revealed that the natural host of the virus may be bats. The study also showed that 2019-nCoV can infect cells with the receptor of SARS-CoV, laying a working foundation for subsequent epidemic prevention and drug research.
Product Recommendation:
没有评论:
发表评论